I doubt it, unless you are asking if mapping would be similarly bad for all centromeres. Here are some repetitive structures (probably not centromeric) that I've found in the nanopore reads for "human" sample NA12878, produced by the nanopore-WGS consortium:
These structures are consistent in that they repeat lots of times, but the internal patterns can be quite different. Here are a few more:

Given that centromeres need to be uniquely linkable to a single chromosome, it would make sense to me if the centromere internal structure is unique to each chromosome.
It's possible to have highly-repetitive structures which don't map to each other. While I haven't dug too deeply into the human reads, I've looked at the 5 most-compressible regions in an assembled rodent parasite (Nippostrongylus brasiliensis) genome, and found no internal similarities between them:
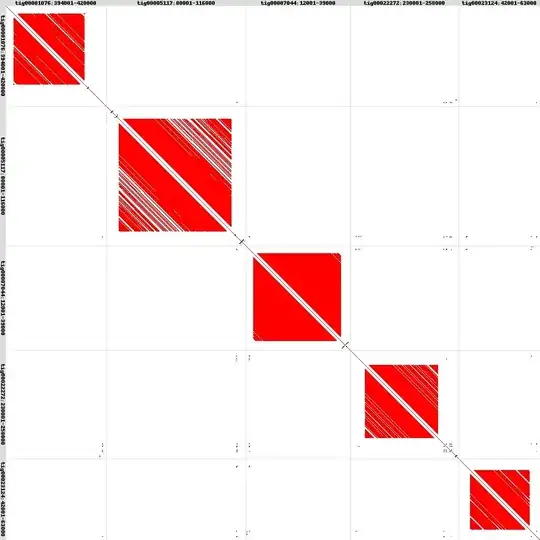
One of the issues with assembly from Illumina reads is that these highly-repetitive regions get collapsed into a single repeat (or at best a region up to twice the fragment length). With internal repeat units that have identity of over 98%, assembling the true sequence is very difficult, even with an exact knowledge of paired read separation. Even if this were possible, it can be impossible to correctly place a read because multiple internal units could be identical (or similarly different) to the sequenced read.


