I have a set of RNA-seq samples from different experiments (Single and Paired End, depending on the experiment). I ran FASTQC in all the samples and found overrepresented adapter sequences: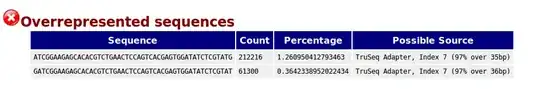
I removed the adaptors (TruSeq adaptors) using Cutadapt (in addition, I removed low quality and N bases from the 3' end of the reads). After that, I ran again FASTQC and the output is the following (representative example) :

Does anyone know what is happening? Now I have an overrepresented sequence for which no sequence is provided. What does this mean?