What does the IGV refseq genes with three different thicknesses of lines mean?
Asked
Active
Viewed 514 times
5
-
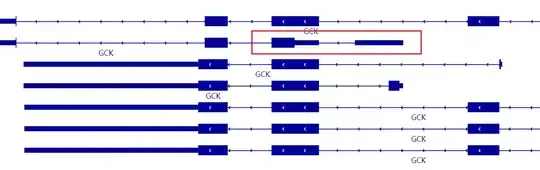
In gene tracks, exons ( regions of the transcript that aligning with the genome ) would be depicted as blocks, while introns would be drawn as the horizontal lines that connect the exons. The direction of transcription would be indicated by arrowheads on the introns. Coding regions of exons would be depicted as tall blocks, while non-coding exons are shorter. The 5'-most exon of each transcript is shorter on the left, indicating an untranslated region, and is taller on the right, indicating a coding sequence. – envs_h_gang_5 Apr 25 '23 at 03:39
1 Answers
5
The thinkness indicates whether something is an intron, a coding exon, or a UTR. The very thin lines are introns, the full thickness boxes are coding exons and the thinner boxes are UTRs.
UTRs, untranslated regions, are exons, they are included in the final mRNA, but they don't code for protein. So, by convention, they are shown as thinner exons to give the user a visual indication that although this is still an exonic region, it's a special one.
I don't really know where this is documented, it's pretty standard across all genome browsers and bam visualizers I know though so it's a common idiom.
terdon
- 10,071
- 5
- 22
- 48