The ladder distance (LD) between residue $i$ and residue $j$ is the minimum number of base pairs (created due to secondary structure of the ssRNA) one has to cross when going from $i$ to $j$.
The Maximum Ladder Distance of an ssRNA molecule is the maximum ladder distance between any two of the residues of this molecule[*].
Given the secondary structure of an ssRNA molecule in dot-bracket notation (e.g. by RNAfold), how can I calculate the Maximum Ladder Distance of this molecule?
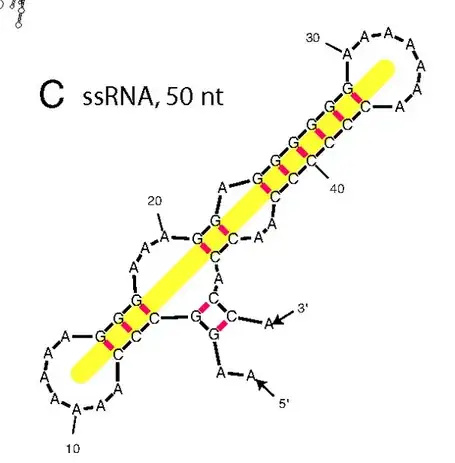
Below is an illustration from [*]. The MLD is 11, since this is the number of basepairs on the yellow line.
The dot-bracket notation for this molecule would be
..(((((.......))...((.((((((.......))))))..)).)).
*: Yoffe AM, Prinsen P, Gopal A, Knobler CM, Gelbart WM, et al. (2008) Predicting the sizes of large RNA molecules. Proc Natl Acad Sci U S A 105: 16153–16158
This question was originally posted in the biology stackexchange: How to calculate the Maximum Ladder Distance (MLD) of ssRNA from Dot-Bracket notation?