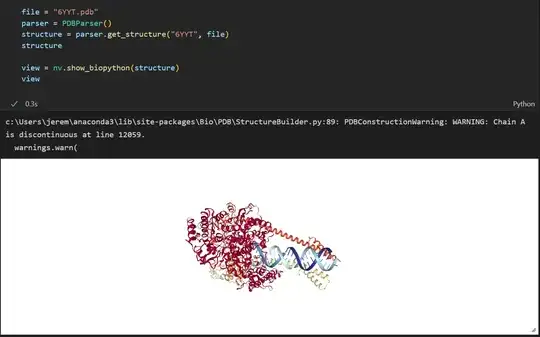
So I have been trying to get into visualizing proteins in python, so after some research I ended up on a tutorial that was teaching you how to visualize a protein from the COVID-19 virus, so I went and setup anaconda, got jupyter notebook working in vscode, and downloaded the necessary files from the PDB database, and made sure they were in the same directory as my notebook but when I run the the nglview.show_biopython(structure) function I get ValueError: I/O opertation on a closed file. I'm stymied this is my first time using jupyter notebook so maybe there is something I'm missing, I don't know.
This what the code looks like
from Bio.PDB import *
import nglview as nv
parser = PDBParser()
structure = parser.get_structure("6YYT", "6YYT.pdb")
view = nv.show_biopython(structure)
The error looks like this
Output exceeds the size limit. Open the full output data in a text editor
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
~\AppData\Local\Temp\ipykernel_8776\2743687014.py in <module>
----> 1 view = nv.show_biopython(structure)
c:\Users\jerem\anaconda3\lib\site-packages\nglview\show.py in show_biopython(entity, kwargs)
450 '''
451 entity = BiopythonStructure(entity)
--> 452 return NGLWidget(entity, kwargs)
453
454
c:\Users\jerem\anaconda3\lib\site-packages\nglview\widget.py in init(self, structure, representations, parameters, kwargs)
243 else:
244 if structure is not None:
--> 245 self.add_structure(structure, kwargs)
246
247 if representations:
c:\Users\jerem\anaconda3\lib\site-packages\nglview\widget.py in add_structure(self, structure, kwargs)
1111 if not isinstance(structure, Structure):
1112 raise ValueError(f'{structure} is not an instance of Structure')
-> 1113 self._load_data(structure, kwargs)
1114 self._ngl_component_ids.append(structure.id)
1115 if self.n_components > 1:
...
--> 200 return io_str.getvalue()
201
202
ValueError: I/O operation on closed file
I only get this error when using nglview.show_biopython, when I run the get_structure() function it appears to read the file just fine. I can visualize other molucles just fine, or maybe that's because I was using the ASE library instead of a file. I don't know, that's why I'm here.
Update: Recently I found out that I can visualize the protein using nglview.show_file() instead of using nglview.show_biopython(). Even though I can visualize proteins now and technically my problem has been solved I would still like to know why the show_biopython() function isn't working properly.
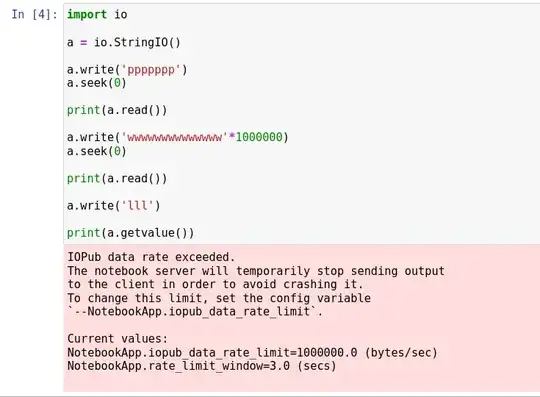
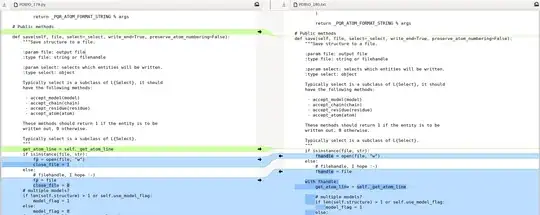
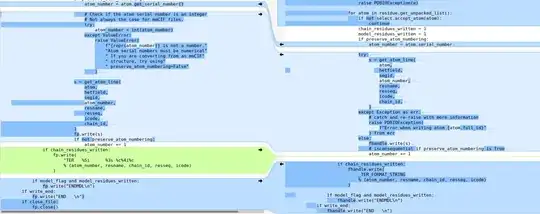
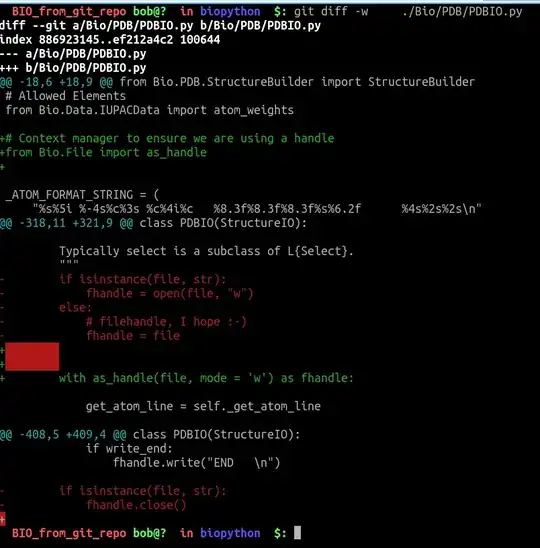
show_biopythonfunction many moons ago. In particular it seems that theStringIOobject at https://github.com/nglviewer/nglview/blob/master/nglview/adaptor.py#L193-L200 gets closed early, preventing it being read. – jgreener Dec 12 '22 at 15:45