I have a set 189 taste receptor protein sequences (not aligned) in a fasta file. I like to get the phylogenetic tree in newick format. I was using earlier https://ngphylogeny.fr/. Unfortunately now it is not working due to some bugs. Necessarily, I just need newick information for the input fasta file.
Any help would be highly appreciated.
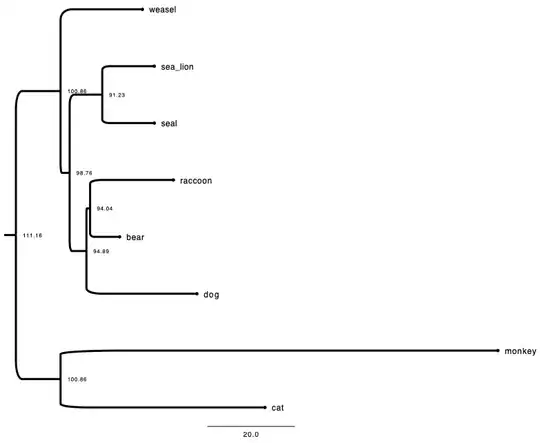
Here is one newick example:
((raccoon:19.19959,bear:6.80041):0.84600,
((sea_lion:11.99700, seal:12.00300):7.52973,
((monkey:100.85930,cat:47.14069):20.59201,
weasel:18.87953):2.09460):3.87382,
dog:25.46154);