I am currently implementing some metrics I could use for comparing two conformations of the same protein in Python.
For example, I know I could use the RMSD of all protein ATOMS in BioPandas using this:
biopandas.pdb.PandasPdb.rmsd(conformation1.df['ATOM'], conformation2.df['ATOM'])
Are there some out-of-the-box tools in Python for measuring the RMSD between the side chains of the two conformations? When trying to do this manually, the main issue is how to identify side chain atoms, which is not clear for me. I know how to get all atoms from a residue using BioPandas:
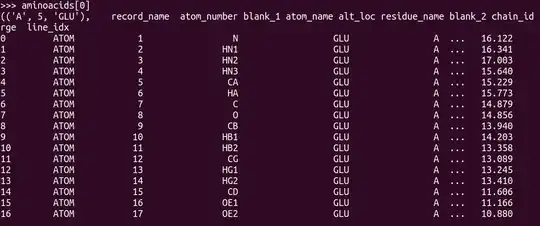
aminoacids = list(PandasPdb().read_pdb(my_pdb_filename).df['ATOM'].groupby(['chain_id', 'residue_number', 'residue_name']))
Now aminoacids[i] is a dataframe containing all the atoms of the i-th aminoacid, like this:
From here, it's not clear how to separate back bone atoms from side chain atoms. It's clear that the one with atom_number 'CA' will be the alpha-Carbon, but I am not sure how to identify the others. But once the separation is done, the side chain RMSD could be measure for example using Bio.SVDSuperimposer.SVDSuperimposer which works directly on the array of 3D coordinates of the atoms.
Thanks!