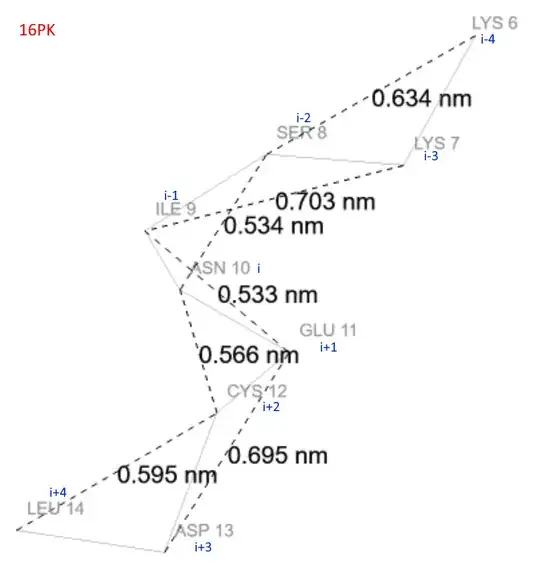
Suppose, we have a protein (16PK).
We are considering a 5-residue segment/window.
If we assume ASN10 to be the ith residue, how can I calculate the following distances in Python?
- from [i+2] to [i-2]
- from [i+1] to [i-2]
- from [i+2] to [i-1]
Next major question is, what would be the distances like if my segment/window is 7 or 9 residues long?