The vertebrate genome project (VGP)
has a lot of interesting publications such as this one.
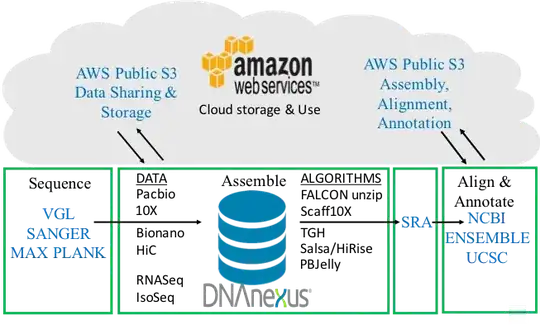
The rough pipeline is outlined below:
Here the pipeline in more detail:
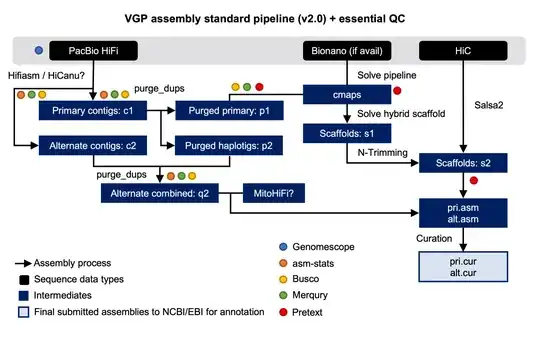
While the paper describes all the steps of the iterative assembly pipeline, I cannot find information on the runtime of their assembly-pipeline.
I was excited to check out their github page to find out more in the documentation. Unfortunately did I not find any information on the runtime in the https://github.com/VGP/vgp-assembly repo.
I'have written some e-mails asking collaborators of VGP but I have still not gotten any answers.
Did any of you apply this assembly-pipeline? Could anyone tell me the runtime of the assembly-pipeline?
I hope this question is in the appropriate stackexchange, if not please point me to what you think fits best. Thanks for your suggestions, please be gentle I'm new to all of this.
For more info on the VGP check out this link