I have a library of biological implementations (i.e. plasmids) of genetic circuits and their experimental data associated. I want to implement an in silico version with SBOL, but to be consistent with what I have I'm my freezer, I need to identify not only the genetic circuit but also the biological replicate (i.e. the different colonies picked), as they have their own associated experimental data.
1 Answers
Support for distinguishing replicates was added in SBOL 2.2, along with other support for linking data throughout the experiment cycle. I will explain this in terms of SBOL 3, which is simpler, then give the SBOL 2 translation at the end.
For SBOL 3, all designs, including those for a plasmid, a strain, etc, are specified in terms of the Component class. This is just a "paper" design, however, and may or may not actually exist. When you have a real physical aliquot that you want to talk about, this is represented by an Implementation object: this is your biological replicate.
The Implementation doesn't really have any content of its own; its nature is defined by linking back to the intended design with a prov:wasDerivedFrom link. If you've verified that what you got is what you intended, you can supplement that with a built link as well.
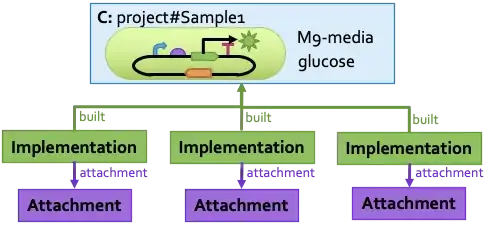
One additional note, however: when you're representing a colony, you're probably not actually representing just a plasmid, but rather are representing a particular strain that has been transformed by a plasmid. The actual data you have may then be one level further removed, since the data has been gathered from a sample under particular growth conditions. SBOL can represent that as well, by describing the sample as a mixture of particular media, inducers, etc., with a transformed strain, which is itself a cell type containing your plasmid.
Here is an example diagram showing just this sort of relationship, of data associated with three replicates of a sample.
You can find this example in the SBOL3 tutorial from COMBINE 2020.
Finally, in SBOL 2, this works exactly the same way, except that instead of a Component your implementation points to a ComponentDefinition or a ModuleDefinition, depending on the nature of the design you're representing. I won't go into that distinction, because it was a bad idea and there's a reason we got rid of it for SBOL 3.
- 653
- 4
- 16
Implementationcan be used in experiments with different media and inducers. – Alejandro Vignoni Mar 16 '21 at 21:17