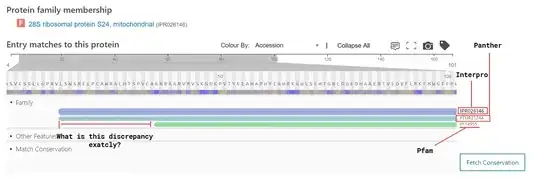
I would be very grateful if somebody could sketch out the methods Pfam and PANTHER use to assign a family to a given protein and how they are different. My (cursory) understanding is that InterpProp pools databases like Pfam and PANTHER and provides a meta-classification based on some combination of their data.
In particular, I wish to understand where the discrepancy comes from described in the picture below?
I'm fairly new to the ecosystem, so feel free to throw any bone this way.