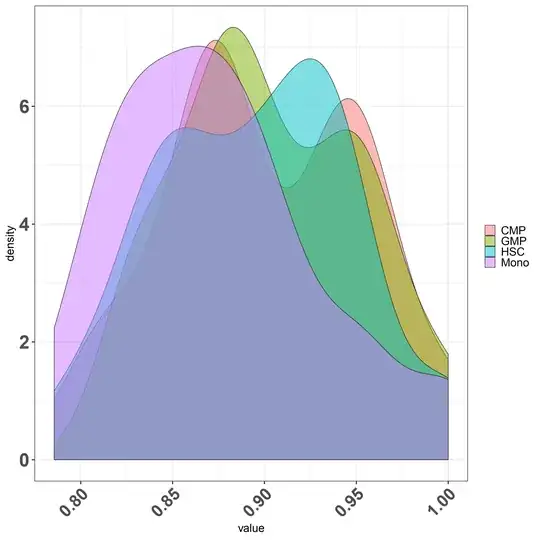
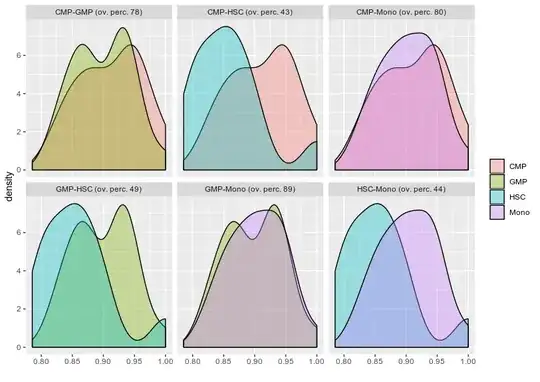
Doubt regarding density plot what is the scale being plotted in the Y axis.Is it possible to scale it to 1?.
This is my code
w <- read.csv("Normal_Myeloid_Dev_stages/Myeloid_non_coding_non_CDS_CORRVALUE.txt",header = TRUE)
head(w)
#names(w)[1] = "Sample"
head(w)
#w.plot <- melt(w)
df <- w
df_melt=melt(df,id.vars="Sample")
#head(df_melt)
tail(df_melt)
df_melt$Group <- gsub('[0-9]', '',df_melt$variable)
head(df_melt)
p1 <- ggplot(aes(x=value, colour=Group), data=df_melt)
p1 + geom_density()
ggplot(df_melt, aes(x = value,y=..density.., fill = Group)) + geom_density(alpha = 0.5)+
theme_bw(base_size=30)+
theme(axis.text.x=element_text(angle = 45, size=45, face="bold", hjust = 1),
axis.text.y=element_text(angle=0, size=50, face="bold", vjust=0.5),
plot.title = element_text(size=40, face="bold"),
legend.title=element_blank(),
legend.key.size=unit(1, "cm"), #Sets overall area/size of the legend
legend.text=element_text(size=30))
dev.off()
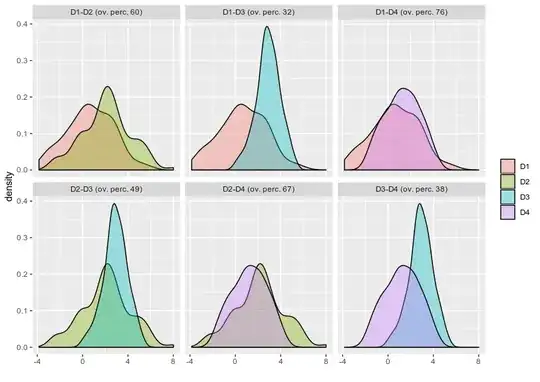
Second question regarding finding the overlapping area between various condition let say I have four condition and i want to find out what is the overlapping or intersecting area between them .
I tried on of the stack solution firstly is it the right way to do it for finding overlap? if yes can i try finding all the condition overlap such as I have HSC,CMP,GMP and Mono.
FDensity = approxfun(density(df_melt$value[df_melt$Group=="CMP"], from=.80, to=1))
MDensity = approxfun(density(df_melt$value[df_melt$Group=="Mono"], from=.80, to=1))
plot(FDensity, xlim=c(.80,1), ylab="Density")
curve(MDensity, add=TRUE)
FminusM = function(x) { FDensity(x) - MDensity(x) }
Intersect = uniroot(FminusM, c(.8, 1))$root
points(Intersect, FDensity(Intersect), pch=20, col="red")
integrate(MDensity, .80,Intersect)$value +
integrate(FDensity, Intersect, 1)$value
Any suggestion or help would be really appreciated
My dataframe small subset
dput(head(df))
structure(list(Sample = structure(c(9L, 10L, 11L, 12L, 1L, 2L
), .Label = c("CMP1", "CMP2", "CMP3", "CMP4", "GMP1", "GMP2",
"GMP3", "GMP4", "HSC1", "HSC2", "HSC3", "HSC4", "Mono1", "Mono2",
"Mono3", "Mono4"), class = "factor"), HSC1 = c(1, 0.901758000194052,
0.880971458703505, 0.900098712568466, 0.873507054094022, 0.906315987662777
), HSC2 = c(0.901758000194052, 1, 0.945122894369186, 0.955453795442752,
0.844777154811663, 0.960260413763721), HSC3 = c(0.880971458703505,
0.945122894369186, 1, 0.931891528389177, 0.829521543809914, 0.93636611655036
), HSC4 = c(0.900098712568466, 0.955453795442752, 0.931891528389177,
1, 0.852155096935692, 0.947192034704188), CMP1 = c(0.873507054094022,
0.844777154811663, 0.829521543809914, 0.852155096935692, 1, 0.88406635204624
), CMP2 = c(0.906315987662777, 0.960260413763721, 0.93636611655036,
0.947192034704188, 0.88406635204624, 1), CMP3 = c(0.883085447599108,
0.928929790213059, 0.937278860333014, 0.923139729112196, 0.870595169861622,
0.958330253497026), CMP4 = c(0.903500691840647, 0.931015449611016,
0.915510499719426, 0.949501343959892, 0.89151869084162, 0.960206335031192
), GMP1 = c(0.859320159223793, 0.833454007185579, 0.821479398953591,
0.837605565046685, 0.856892209387879, 0.8612685942226), GMP2 = c(0.894680165774456,
0.944129985346879, 0.92461599550388, 0.937108867595306, 0.875930650641323,
0.966512850930959), GMP3 = c(0.881416749993221, 0.925001567707159,
0.923015420025178, 0.920758302664238, 0.863998116268766, 0.95069438729762
), GMP4 = c(0.887230702792805, 0.911614382740949, 0.898704725226052,
0.938310378760322, 0.875956580458754, 0.93865124308053), Mono1 = c(0.80834274546097,
0.803706049167148, 0.785535924609134, 0.794438848049474, 0.794402126200663,
0.810382054101301), Mono2 = c(0.829928035079747, 0.868021129791583,
0.848929312833536, 0.852953184941131, 0.818074585324121, 0.880490870699251
), Mono3 = c(0.834028389107401, 0.859818911060746, 0.852694253707916,
0.85039434449057, 0.82251614206182, 0.872740655210769), Mono4 = c(0.832524159512094,
0.858444209302146, 0.84397886802844, 0.862850406374668, 0.818436260136434,
0.870293804485003)), row.names = c(NA, 6L), class = "data.frame")
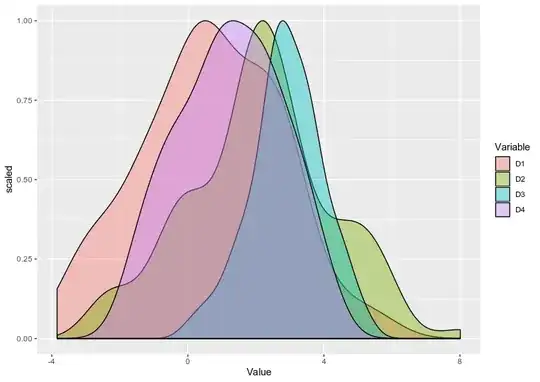
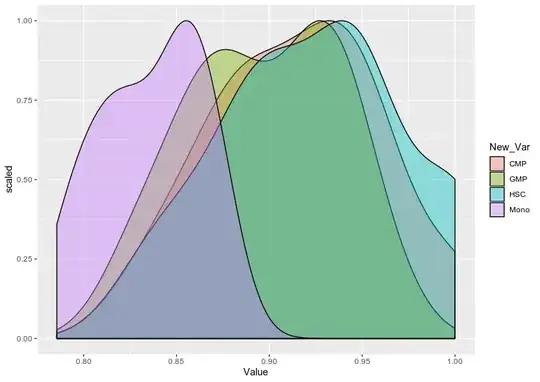
scaledfunction is really required for that. – dc37 Dec 17 '19 at 22:23