Downloading entire genomes
The best way to download FASTA sequences for an entire genome is to search for the genome, for example Theobroma cacao genome in the NCBI Assembly portal and use the big blue Download button.
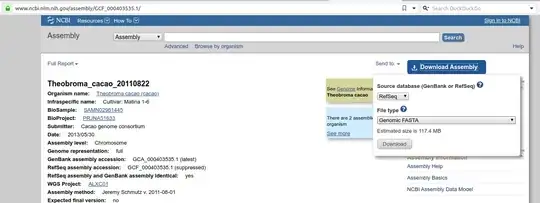
Downloading individual chromosomes
For a given assembly, if you want to download the FASTA sequences for a bunch of chromosomes, you can do that by going to the Genomes FTP path highlighted in the screenshot:
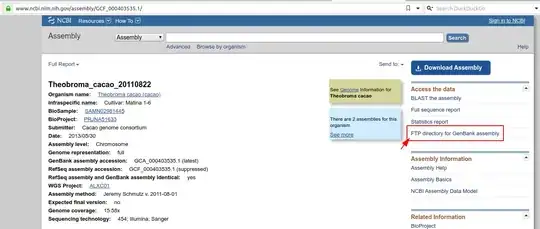 Once you are in the Genomes FTP path, you can navigate to the FASTA folder as follows:
Theobroma FTP directory >> Assembly Structure >> Primary Assembly >> Assembled Chromosomes >> FASTA.
Once you are in the Genomes FTP path, you can navigate to the FASTA folder as follows:
Theobroma FTP directory >> Assembly Structure >> Primary Assembly >> Assembled Chromosomes >> FASTA.
Downloading a few sequences
For this, you can use Entrez Direct as mentioned by @dc37. Why not always use Entrez Direct? While it is fine for a small number of sequences, it can be slow to download a large number of sequences. Entrez Direct by default will download uncompressed data so you will end up spending more time downloading a larger file instead of downloading a smaller, compressed file from FTP more quickly.
If you were to use Entrez Direct for this purpose, I'd not bother with a bash script and use epost to first post the entire list of accessions and then pipe it to efetch as shown below:
epost -db nuccore -input <file_with_accs> -format acc \
| efetch -format fasta > sequences.fasta


NCBI_API_KEYfor batch downloading withGNU parallelat a max rate of 10 sequences/s. Then save accessions in one file and runparallel -a accession.txt -j8 'efetch -db nuccore -id {} -format fasta >{}.fa'. Adjust-j8to-j3if you do not have a key. – Forrest Vigor Dec 06 '22 at 07:57